Kai Zhu 「祝凯」

I am a Ph.D. student in Pharmaceutics at Zhejiang University, advised by Prof. Hou Tingjun. I am also a visiting student at the Italian Institute of Technology, working with Luigi Bonati and Prof. Michele Parrinello.
My research focuses on developing machine learning–based enhanced sampling methods and their applications to atomic systems.
I enjoy running 🏃♂️, boxing 🥊, and swimming 🏊♂️; my favorite athlete is Georges St‑Pierre.
Research Interests
- Atomistic simulations
- Enhanced sampling
- Machine learning
- Drug design
Email: 22319143@zju.edu.cn
Publications
† Equal contribution
Machine Learning Enhanced Sampling

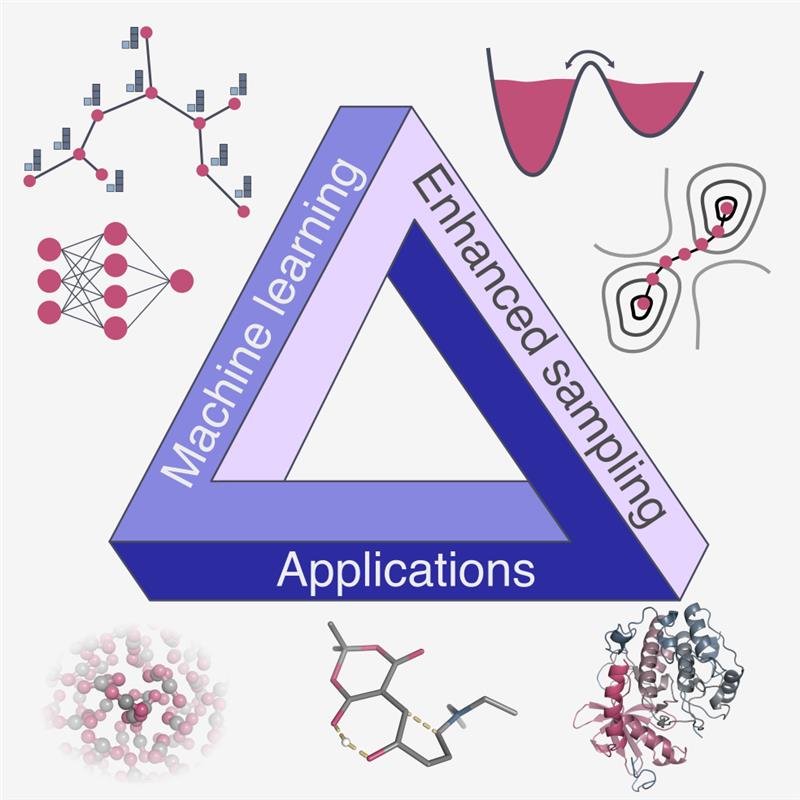
Molecular dynamics simulations hold great promise for providing insight into the microscopic behavior of complex molecular systems. However, their effectiveness is often constrained by long timescales associated with rare events. Enhanced sampling methods have been developed to address these challenges, and recent years have seen a growing integration with machine learning techniques. This Review provides a comprehensive overview of how they are reshaping the field, with a particular focus on the data-driven construction of collective variables. Furthermore, these techniques have also improved biasing schemes and unlocked novel strategies via reinforcement learning and generative approaches. In addition to methodological advances, we highlight applications spanning different areas, such as biomolecular processes, ligand binding, catalytic reactions, and phase transitions. We conclude by outlining future directions aimed at enabling more automated strategies for rareevent sampling.
Self‑Supervised Evolution Operator Learning for High‑Dimensional Dynamical Systems
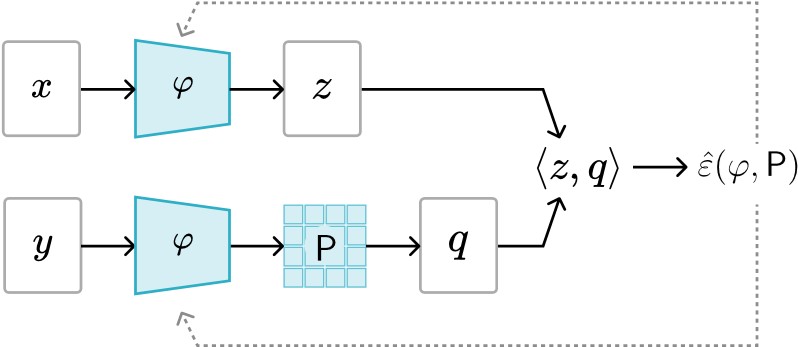
We introduce an encoder-only approach to learn the evolution operators of large-scale non-linear dynamical systems, such as those describing complex natural phenomena. Evolution operators are particularly well-suited for analyzing systems that exhibit complex spatio-temporal patterns and have become a key analytical tool across various scientific communities. As terabyte-scale weather datasets and simulation tools capable of running millions of molecular dynamics steps per day are becoming commodities, our approach provides an effective tool to make sense of them from a data-driven perspective. The core of it lies in a remarkable connection between self-supervised representation learning methods and the recently established learning theory of evolution operators. To show the usefulness of the proposed method, we test it across multiple scientific domains: explaining the folding dynamics of small proteins, the binding process of drug-like molecules in host sites, and autonomously finding patterns in climate data. Code and data to reproduce the experiments are made available open source.
Machine Learning Force Fields
A Scalable and Quantum‑Accurate Foundation Model for Biomolecular Force Field via Linearly Tensorized Quadrangle Attention
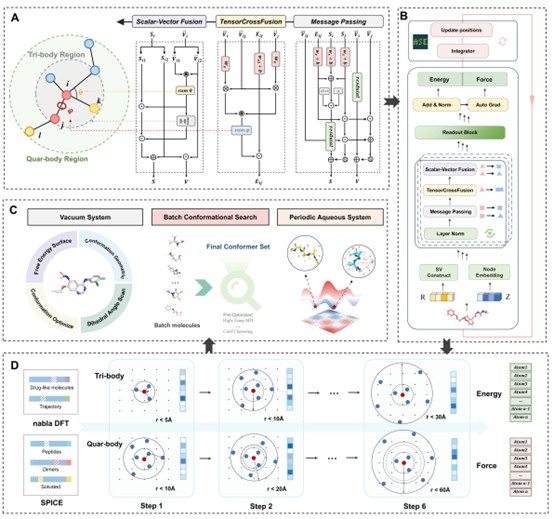
Accurate atomistic biomolecular simulations are vital for disease mechanism understanding, drug discovery, and biomaterial design, but existing simulation methods exhibit significant limitations. Classical force fields are efficient but lack accuracy for transition states and fine conformational details critical in many chemical and biological processes. Quantum Mechanics (QM) methods are highly accurate but computationally infeasible for large-scale or long-time simulations. AI-based force fields (AIFFs) aim to achieve QM-level accuracy with efficiency but struggle to balance many-body modeling complexity, accuracy, and speed, often constrained by limited training data and insufficient validation for generalizability. To overcome these challenges, we introduce LiTEN, a novel equivariant neural network with Tensorized Quadrangle Attention (TQA). TQA efficiently models three- and four-body interactions with linear complexity by reparameterizing high-order tensor features via vector operations, avoiding costly spherical harmonics. Building on LiTEN, LiTEN-FF is a robust AIFF foundation model, pre-trained on the extensive nablaDFT dataset for broad chemical generalization and fine-tuned on SPICE for accurate solvated system simulations. LiTEN achieves state-of-the-art (SOTA) performance across most evaluation subsets of rMD17, MD22, and Chignolin, outperforming leading models such as MACE, NequIP, and EquiFormer. LiTEN-FF enables the most comprehensive suite of downstream biomolecular modeling tasks to date, including QM-level conformer searches, geometry optimization, and free energy surface construction, while offering 10x faster inference than MACE-OFF for large biomolecules (~1000 atoms). In summary, we present a physically grounded, highly efficient framework that advances complex biomolecular modeling, providing a versatile foundation for drug discovery and related applications.
Machine Learning Drug Design

Revisiting Protein–Protein Docking: A Systematic Evaluation Framework
Protein–protein interactions play pivotal roles in a wide range of biological processes. Determining the atomic-level structures of protein–protein complexes is indispensable for elucidating macromolecular interaction mechanisms and advancing structure-based drug design. Protein–protein docking, as one of the leading computational approaches for predicting complex structures, has seen considerable progress but requires rigorous evaluation in practical applications. In this study, we proposed a comprehensive benchmarking framework to evaluate 11 docking methods spanning traditional (HDOCK, PatchDock, PIPER, ZDOCK) and deep learning (DL)-based (EquiDock, ElliDock, EBMDock, GeoDock, DiffDock-PP, AlphaFold-Multimer, AlphaFold3) approaches. Our framework incorporates the classical DockingBenchmark 5.5 data set for evaluating flexible docking, introduces a newly curated data set (AACBench) for antibody–antigen complex docking, and establishes the PPCBench data set to examine the out-of-distribution (OOD) generalization capabilities of DL-based methods. In docking against apo structures, AlphaFold3 achieves a superior top-5 success rate of 77.98%, whereas the traditional approach HDOCK reaches merely 12.84%, despite its highest top-5 success rate of 85.24% when docking against holo structures. For antibody–antigen docking, AlphaFold3 remains the most accurate method (top-5 success rate: 31.78%) and substantially outperforms AlphaFold-Multimer in modeling the CDR-H3 loop. In OOD generalization tests, all DL-based models exhibit markedly reduced performance on the PPCBench data set. Overall, our work establishes a unified benchmarking framework that enables systematic evaluation of docking methods across diverse tasks and provides critical insights into the strengths and limitations of current docking strategies, thereby informing future developments in protein–protein docking research.
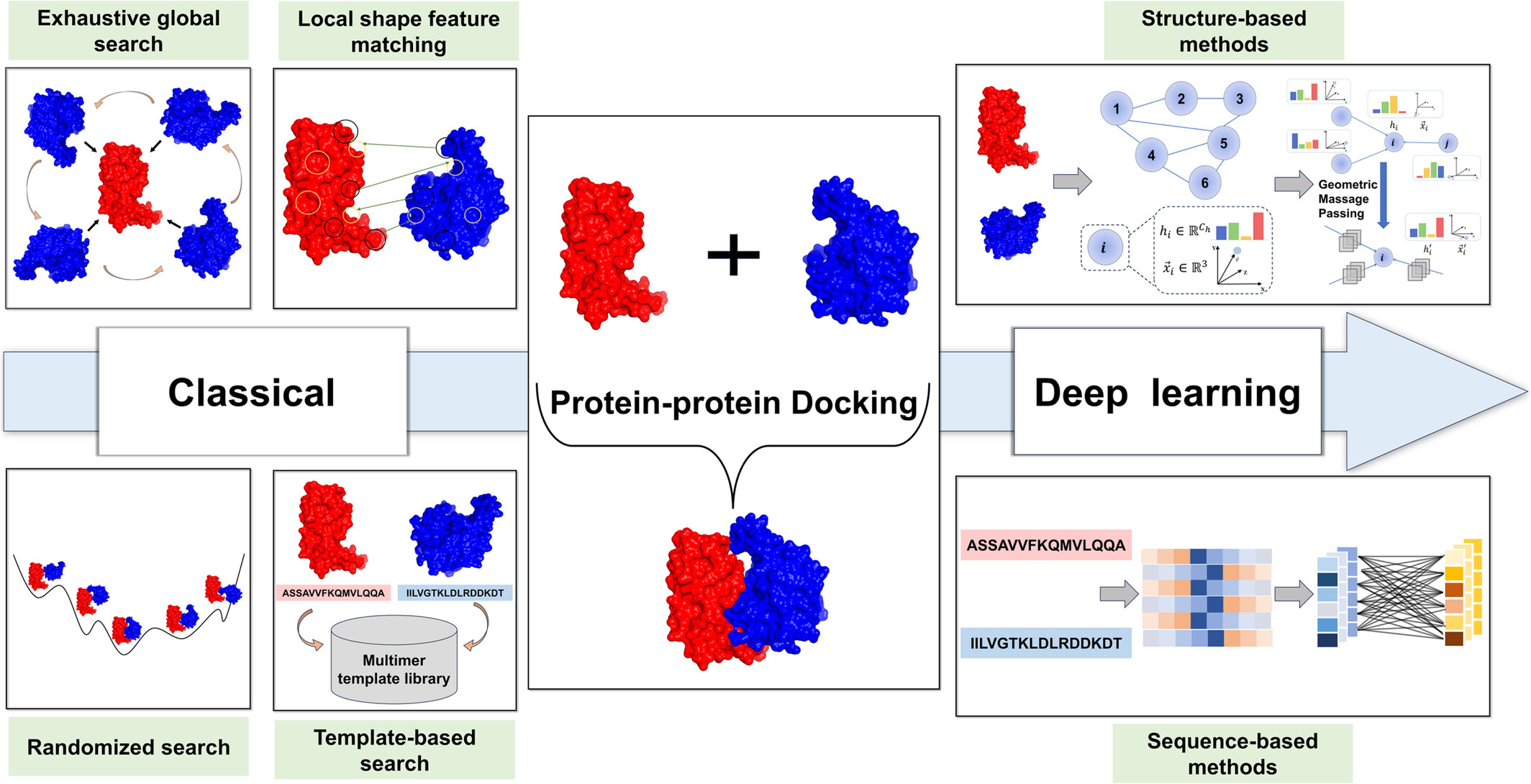
From Traditional Methods to Deep Learning Approaches: Advances in Protein–Protein Docking
Protein–protein interactions play a crucial role in human biological processes, and deciphering their structural information and interaction patterns is essential for drug development. The high costs of experimental structure determination have brought computational protein–protein docking methods into the spotlight. Traditional docking algorithms, which hinge on a sampling‑scoring framework, heavily rely on extensive sampling of candidate poses and customized scoring functions based on the geometric and chemical compatibility between proteins. However, these methods face challenges related to sampling efficiency and stability. The advent of deep learning (DL) has ushered in data‑driven docking methods that demonstrate significant advantages, particularly boosting the efficiency of protein–protein docking. We systematically review the historical development of protein–protein docking from traditional approaches to DL techniques and provide insights into emerging technologies in this field. Moreover, we summarize the commonly used datasets and evaluation metrics in protein–protein docking. We expect that this review can offer valuable guidance for the development of more efficient protein–protein docking algorithms.